Table of Contents
- Browsing proteins
- Editing proteins
- Protein identity check
Editing proteins
You may work with protein text in either of the PLN, UniProt or GPMAW formats. Proteax will automatically recognize the format of the protein text that you enter or copy/paste into the text field in the middle.
To the right you will see the sequence of the protein visualized along with information about the total number of residues and the protein structure's average molecular weight.
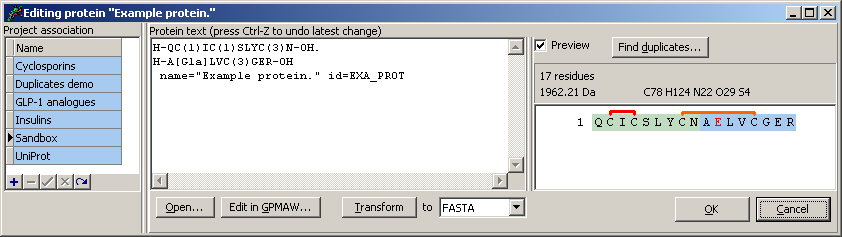
Projects
Each protein entry is asscoiated with a project in order to group related entries. The total list of projects is displayed to the left and the project that the current entry belongs to will be highlighted. You may move the protein entry to a different project by simply clicking that project. Clicking a project name also allows you to rename the project - but only if the project is not locked. Locked projects are displayed with a blue background while open projects have a white background.
You can add and delete projects by using the small buttons below the project list.
A quick PLN primer
The easiest way to enter protein entries by hand is via PLN: Protein Line Notaton. This section will give you a quick introduction to building a protein structure using PLN.
A basic two-chain protein
Let's start by entering a simple two-chain protein. Chains consist in their simplest form of a string of one-letter residue codes. Chains are separated by periods and must be terminated properly.
H-GIVEQCCTSICSLYQN-OH.H-FVNQHLCGSHLFYTPKT-OH
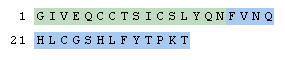
Adding disulfide bonds
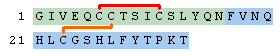
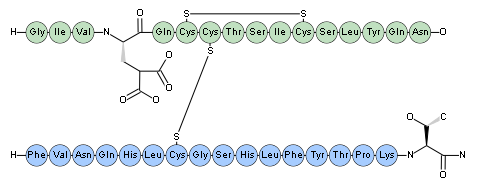
The protein above is really two separate peptide fragments so we should add disulfide bonds to link them together. Cysteines that form disulifde bonds are linked by adding a unique number in parenthesis after the cysteine. The unique number identifies the particular disulfide bond. After adding two disulfide bonds, the PLN becomes the following (Human Insulin with a number of amino acids deleted).
H-GIVEQC(1)C(2)TSIC(1)SLYQN-OH.H-FVNQHLC(2)GSHLFYTPKT-OH
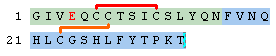
Finally, we can add a modified residue and a terminal modification. Glu-4 is changed into "Gla" aka. "4-carboxyglutamate" and the second chain is terminated with an amidated residue.
H-GIV[Gla]QC(1)C(2)TSIC(1)SLYQN-OH.H-FVNQHLC(2)GSHLFYTPKT-[NH2]
Cyclic chains
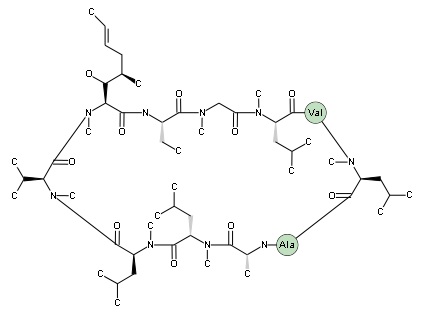
If you have a fully cyclic chain, specify the terminals as "(cyclo)", like below where cyclosporin CsA is defined. The sequence contains a D-form alanine; D-forms are created by prefixing a residue with "{d}".
(cyclo)-[MeBmt][Abu][MeGly][MeLeu]V[MeLeu]A{d}A[MeLeu][MeLeu][MeVal]-(cyclo)

Lactam cyclizations
Nitro groups and acid groups can be crosslinked by using the "(lactam)" or "(cyclo)" keywords.
You must add a unique number after "lactam" or "cyclo" to tell Proteax which groups to link. Proteax will automatically detect which end of the crosslink has the nitro group and which end has the acid group. Here is an example of sidechain-sidechain cyclization within a peptide chain.
H-ADK(cyclo1)ED(cyclo1)FG-OH
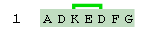
You can also create terminal-sidechain crosslinks, simply by placing the "(cyclo)" keyword at a terminal, like shown below.
Note that the unique number after "cyclo" is required when "(cyclo)" links from a terminal to a sidechain. It is only when "(cyclo)" is used to specify a fully cyclic chain that you may omit the unique number.
(cyclo1)-ADKED(cyclo1)FG-OH

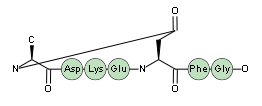
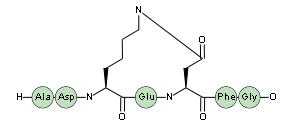
Sidechain-terminal cyclizations can also be used to form branched peptides.
H-ASD(lactam1)EFG-OH.(lactam1)-QWEKY-OH

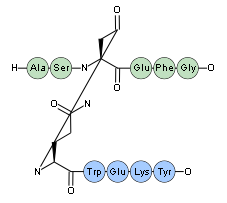
Note that the "(lactam)" keyword will be read as a synonym for "(cyclo)" - you may use them interchangably as you wish. However, when Proteax outputs PLN it will always use "(cyclo)" for in-chain cyclizations and "(lactam)" for inter-chain crosslinks.
This concludes the brief overview of PLN. If you have any questions or topics that you feel have been inadequately adressed please feel free to .

