Proteax Cartridge - protein chemistry for Oracle® databases
Add protein support to your database
Proteax Cartridge lets you work with chemically or post-translationally modified protein sequences in Oracle databases via normal SQL statements.
|
![A simple SQL statement that calculates the average molecular weight of the GLP-1 molecule. [Image: A simple SQL statement that calculates the average molecular weight of the GLP-1 molecule.]](sql_glp-1_avg_mw.png)
Average molecular weight of the GLP-1 molecule calculated with a simple SQL statement. |
Industry standard formats
Proteax Cartridge accepts protein entries in the following standard protein file formats:
Protein entries can be freely converted to any of the formats PLN, UniProt, and GPMAW without loss of chemical information. Converting entries to FASTA format will by its very nature strip all chemical annotations. |
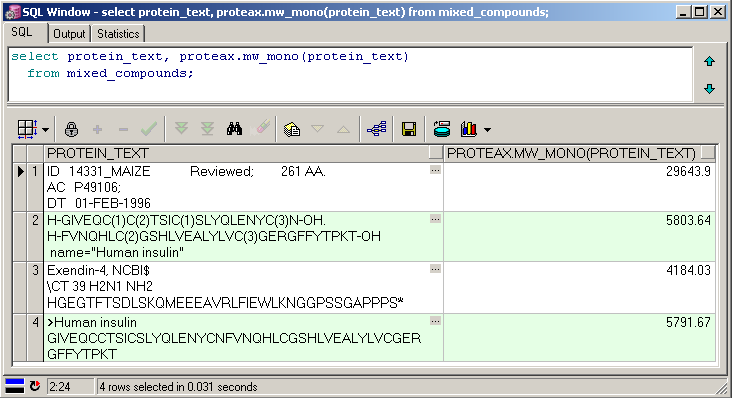
Any supported input format is accepted. |
Functional indexing
Proteax Cartridge provides a number of functions that can be indexed by standard Oracle commands. By leveraging Oracle's advanced support for functional indexes seamless integration into the Oracle optimizer is ensured.
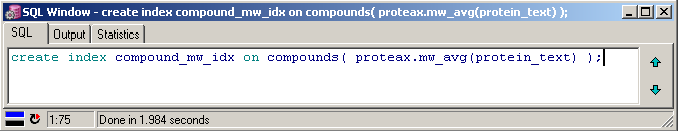
Create protein-based indexes through normal Oracle commands. |
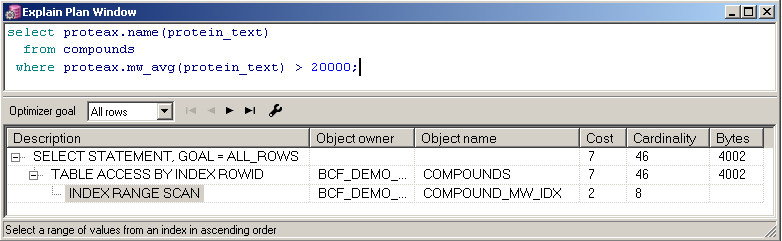
Indexed Proteax functions are utilized as expected by the Oracle cost-based optimizer. |
Protein comparison
Proteax Cartridge contains powerful normalization functions that can be used to determine the identity of two protein entries even in the presence of cyclic chains. Normalization can be done to check sequence identity or to obtain a good measure of structural identity. The normalization functions can be indexed to ensure excellent performance of identity checks.
From Q1 2010 the Proteax Cartridge will support DerNot expressions. DerNot expressions specify edit operations - residue substitutions, insertions, deletions, terminal modifications, and chain extensions. By applying DerNot expressions you can create new derivatives from existing entries or you can let Proteax calculate the DerNot expressions that describe the difference between similar proteins.
Open Source visualization tools
|
Sequence visualization tools can be downloaded for free from the downloads area. The tools are released with source code under a BSD-style license which lets you adapt and translate the tools to your environment of choice. Note that the tools are only hundreds of lines of code, not thousands. Proteax Cartridge is able to generate the rendering info required by the visualization tools. The Proteax demo site shows real-life examples of the usage of the visualization tools. |
![Flash-based sequence rendering on the web. [Image: Flash-based sequence rendering on the web.]](flash_sequence_rendering.png)
Part of the Proteax Live demo web page showing sequence graphics created by Biochemfusion's Flash rendering tool. |
Chemical indexing
Proteax Cartridge can effectively extend the usable molecular size range of existing chemistry cartridges. Chemistry databases will readily index small peptides but will normally not scale well to large proteins.
Proteax Cartridge can produce condensed representation molecules - a format that enhances registration and search performance by orders of magnitude. This will allow your chemistry database to store and chemically search protein structures with thousands of residues.
Proteax Cartridge generates condensed chemical structures that adhere to the rules defined in the article "Building a BioChemformatics Database", Jan H. Jensen, Thomas Hoeg-Jensen and Søren B. Padkjær, J. Chem. Inf. Model., November 2008.
System requirements
Proteax Cartridge installs on servers running a 32- or 64-bit version of Linux® or Windows®. In order to install Proteax Cartridge you will also need to have an existing Oracle 10g or 11g database running on your server. The following Oracle configurations are supported.
| Oracle 10gR2 | Oracle 11gR1 | Oracle XE (10g) | ||
|---|---|---|---|---|
| Windows |
32-bit 64-bit |
32-bit 64-bit | 32-bit | |
| Linux |
32-bit 64-bit |
32-bit 64-bit | 32-bit |
Licensing
Commercial licensing details are available by contacting . Educational and non-profit research institutions may apply for free academic licenses by contacting .


![Full-structure 2D chemical structure generation. [Image: Full-structure 2D chemical structure generation.]](mol_full_structure.png)
![Condensed 2D chemical structure generation. [Image: Condensed 2D chemical structure generation.]](mol_condensed.png)